DNN with backpropagation in C++, part 5
Adding Softmax layer
In this post I’ll modify the previous example adding Softmax layer.
Forward path for the layer is Softmax function
Softmax function produces M-dimentional vector output \(\sigma {(\boldsymbol{x})}\) for M-dimentional input vector X, and is defined as
\[\sigma {(\boldsymbol{x})_{i}} = \frac {e^{x_{i}}} {\sum_{i=1}^{M} e^{x_{i}}}\]where X is input vector of size M:
\[X = \left( \begin{array}{ccc} x_{1} & x_{2} & \ldots & x_{M} \\ \end{array} \right)\]Plot of \(\sigma {(x_{1})}\)
\[\sigma {(x_{1})} = \frac {e^{x_{1}}} {e^{x_{1}} + e^{x_{2}}}\]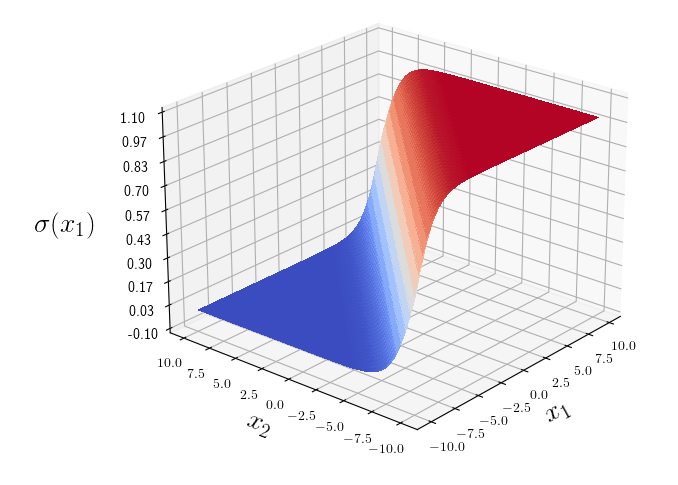
Let’s find Softmax derivative required for backward path
Derivative of a vector function y(x) with respect to input vector x, is a Jacobian matrix.
Derivative of softmax function \(\frac {\partial {\sigma}} {\partial x}\) is a Jacobian of first order partial derivatives:
\[J = \left( \begin{array}{ccc} \frac {\partial {\sigma_{1}}} {\partial x_{1}} & \frac {\partial {\sigma_{1}}} {\partial x_{2}} & \ldots & \frac {\partial {\sigma_{1}}} {\partial x_{M}} \\ \frac {\partial {\sigma_{2}}} {\partial x_{1}} & \frac {\partial {\sigma_{2}}} {\partial x_{2}} & \ldots & \frac {\partial {\sigma_{2}}} {\partial x_{M}} \\ \vdots & \vdots & \ldots & \vdots \\ \frac {\partial {\sigma_{M}}} {\partial x_{1}} & \frac {\partial {\sigma_{M}}} {\partial x_{2}} & \ldots & \frac {\partial {\sigma_{M}}} {\partial x_{M}} \\ \end{array} \right)\]Let’s find \(\frac {\partial {\sigma_{i}}} {\partial x_{i}}\)
Making
\[\sigma {(\boldsymbol{x})_{i}} = \frac {g_{i}} {h(x)}\]where
\[g_{i} = e^{x_{i}} \\ h(x) = {\sum_{i=1}^{M} e^{x_{i}}}\]Taking derivative
\[\frac {\partial {\sigma_{i}}} {\partial x_{j}} = \frac {\frac {\partial {g_{i}}} {\partial {x_{j}}} * h - \frac {\partial {h}} {\partial {x_{j}}} * g_{i}} {h^2}\]Wehere
\[\frac {\partial {h}} {\partial {x_{j}}} = e^{x_{j}}\]And
\[\frac {\partial {g_{i}}} {\partial {x_{j}}} = \begin{cases} e^{x_{j}}, i = j \\ 0, i \ne j \end{cases}\]Using Kroneker delta for \(\frac {\partial {g_{i}}} {\partial {x_{j}}}\)
\[\delta_{ij} = \begin{cases} 0, i \ne j \\ 1, i = j \end{cases}\] \[\frac {\partial {g_{i}}} {\partial {x_{j}}} = \delta_{ij} * e^{x_{j}}\]We get:
\[\frac {\partial {\sigma_{i}}} {\partial x_{j}} = \frac {\delta_{ij} * e^{x_{j}} * h - e^{x_{j}} * e^{x_{i}}} {h^2}\]Simplifying further
\[\frac {\partial {\sigma_{i}}} {\partial x_{j}} = \frac {e^{x_{j}}} {h} * \frac {\delta_{ij} * h - e^{x_{i}}} {h}\]We get final expression for Softmax derivative
\[\frac {\partial {\sigma_{i}}} {\partial x_{j}} = \sigma_{j} * ({\delta_{ij} - \sigma_{i}})\]Putting it into Jacobian:
\[J = \left( \begin{array}{ccc} \sigma_{1}*(1 - \sigma_{1}) & -\sigma_{1} * \sigma_{2} & \ldots & - \sigma_{1} * \sigma_{M} \\ -\sigma_{2}*\sigma_{1} & \sigma_{2}*(1 - \sigma_{2}) & \ldots & -\sigma_{2}*\sigma_{M} \\ \vdots & \vdots & \ldots & \vdots \\ -\sigma_{M}*\sigma_{1} & -\sigma_{M}*\sigma_{2} & \ldots & - \sigma_{M}*(1 - \sigma_{M}) \\ \end{array} \right)\]Assuming loss function is Mean Squared Error
\[E(Y) = MSE(Y, \hat Y) = \frac {1} {N} \sum_{i=0}^{N-1} ( Y_{i} - \hat Y_{i} )^2\]Softmax gradient backpropagation output is dot product of MSE derivative and Jacobian J:
\[y = \sigma {(\boldsymbol{x})} \\ E(y) = E(\sigma {(\boldsymbol{x})}) \\ \frac {\partial E} {\partial x} = \frac {\partial E} {\partial y} * \frac {\partial y} {\partial x}\] \[\frac {\partial E} {\partial x} = \frac {2} {N} (\hat {Y} - Y) * J\]Finally, let’s write some code
First I’ll modify python code from previous example to use bias in dense layers.
For this experiment I’ve used the following software versions:
$ python3 -m pip freeze | grep "numpy\|tensorflow"
numpy==1.19.5
tensorflow==2.5.0rc2
$ g++ --version
g++ 9.3.0Python code will be very similar to Python sample from the prevoius post. I’ll disable activation for the 2nd dense layer and add Softmax as the final DNN layer
import tensorflow as tf
from tensorflow.keras.layers import Dense
from tensorflow.keras.optimizers import RMSprop
import numpy as np
num_inputs = 2
num_outputs = 2Create 3 layer sequential model
num_inputs = 2
num_outputs = 2
# Create sequential model
model = tf.keras.Sequential()
layer1 = Dense(units=num_inputs, use_bias=True, activation="sigmoid", weights=[np.ones([num_inputs, num_inputs]), np.full([num_inputs], 2.0)])
layer2 = Dense(units=num_outputs, use_bias=True, activation=None, weights=[np.array([[1.0, 2.0], [3.0, 2.0]]), np.array([1.0, 2.0])])
layer3 = Softmax(axis=-1)
model.add(layer1)
model.add(layer2)
model.add(layer3)Use mean square error for the loss function.
# use MSE as loss function
loss_fn = tf.keras.losses.MeanSquaredError()Hardcode model input and expected model output. We’ll use the same array values later in C++ implementation.
# Arbitrary model input
x = np.array([-1.0, 0.0])
# Expected output
y_true = np.array([1.0, 0.0])Use Stochastic Gradient Decent (SGD) optimizer.
SGD weight update rule is \(W = W - LR * \nabla\)
\(\nabla\) is weight gradient and \(LR\) is learning rate.
For now we’ll assume learning rate equal to 1.0
# SGD update rule for parameter w with gradient g when momentum is 0 is as follows:
# w = w - learning_rate * g
#
# For simplicity make learning_rate=1.0
optimizer = tf.keras.optimizers.SGD(learning_rate=1.0, momentum=0.0)In the training loop we’ll compute model output for input X, compute and backpropagate the loss.
# Get model output y for input x, compute loss, and record gradients
with tf.GradientTape(persistent=True) as tape:
# get model output y for input x
# add newaxis for batch size of 1
xt = tf.convert_to_tensor(x[np.newaxis, ...])
tape.watch(xt)
y = model(xt)
# obtain MSE loss
loss = loss_fn(y_true, y)
# loss gradient with respect to loss input y
dy_dw = tape.gradient(y, model.trainable_variables)
# loss gradient with respect to loss input y
dloss_dy = tape.gradient(loss, y)
dloss_dw = tape.gradient(loss, model.trainable_variables)
# adjust Dense layer weights
grad = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(grad, model.trainable_variables))Finally we’ll print inputs, outputs, gradients, and updated Dense layer weights.
# print model input and output excluding batch dimention
print(f"input x={x}")
print(f"output y={y[0]}")
print(f"expected output y_true={y_true}")
# print MSE loss
print(f"loss={loss}")
# print loss gradients
print("dloss_dy={}".format(*[v.numpy() for v in dloss_dy]))
# print updated dense layer weights
print("updated weights=")
for idx, layer_vars in enumerate(model.trainable_variables):
print(f"{idx+1}) {layer_vars.name}\n{layer_vars.numpy()}")After running Python example:
$ python3 dense5.py
input x=[-1. 0.]
output y=[0.26894143 0.7310586 ]
expected output y_true=[1. 0.]
loss=0.534446656703949
dloss_dy=[-0.7310586 0.7310586]
updated weights=
1) dense/kernel:0
[[1.05652 0.94348]
[1. 1. ]]
2) dense/bias:0
[1.94348 2.05652]
3) dense_1/kernel:0
[[1.2101572 1.7898428]
[3.2101572 1.7898428]]
4) dense_1/bias:0
[1.2874696 1.7125304]Output of Python dense3.py will be used to validate the following C++ code. Let’s code the same example in C++ Include c++ headers for std containers, math and output.
#include <cstdio>
#include <vector>
#include <algorithm>
#include <cassert>
#include <numeric>
#include <array>
#include <chrono>
#include <iostream>
#include <string>
#include <functional>
#include <array>
#include <iterator>
#include <cmath>
using namespace std;
using std::chrono::high_resolution_clock;
using std::chrono::duration_cast;
using std::chrono::microseconds;Lambda will be used to pretty print inputs, outputs, and layer weights.
/*
* Print helper function
*/
auto print_fn = [](const float& x) -> void {printf("%.1f ", x);};Dense layer weights initializers, adding few more constant initializers as compared to previous example.
/*
* Constant weight intializer
*/
const float const_zero = 0.0;
const float const_one = 1.0;
const float const_two = 2.0;
template<float const& value = const_one>
constexpr auto const_initializer = []() -> float
{
return value;
};Dense layer class template includes forward() and backward() functions. Now there’s few more options to initialize weights and bias values from supplied arrays, similar to how it’s done in Python example above.
/*
* Dense layer class template
*
* Parameters:
* num_inputs: number of inputs to Dense layer
* num_outputs: number of Dense layer outputs
* T: input, output, and weights type in the dense layer
* initializer: weights initializer function
*/
template<size_t num_inputs, size_t num_outputs, typename T = float,
T (*weights_initializer)() = const_initializer<const_zero>,
T (*bias_initializer)() = const_initializer<const_zero> >
struct Dense
{
typedef array<T, num_inputs> input_vector;
typedef array<T, num_outputs> output_vector;
typedef T (*initializer)();
vector<input_vector> weights;
output_vector bias;
bool use_bias = true;
/*
* Default dense layer constructor
*/
Dense(bool _use_bias)
{
/*
* Create num_outputs x num_inputs weights matrix
*/
weights.resize(num_outputs);
for (input_vector& w: weights)
{
generate(w.begin(), w.end(), *weights_initializer);
}
/*
* Initialize bias vector
*/
use_bias = _use_bias;
generate(bias.begin(), bias.end(), *bias_initializer);
}
/*
* Dense layer constructor from provided weigths and biases
* Note: weights are stored transposed
*/
Dense(const array<array<T, num_inputs>, num_outputs>& weights_init,
const array<T, num_outputs>& biases_init)
{
/*
* Create num_outputs x num_inputs weights matrix
*/
for (auto weights_row: weights_init)
{
weights.push_back(weights_row);
}
/*
* Initialize bias vector
*/
bias = biases_init;
}
/*
* Dense layer constructor from provided weigths. Bias is not used
* Note: weights are stored transposed
*/
Dense(const array<array<T, num_inputs>, num_outputs>& weights_init)
{
/*
* Create num_outputs x num_inputs weights matrix
*/
for (auto weights_row: weights_init)
{
weights.push_back(weights_row);
}
use_bias = false;
}
/*
* Dense layer forward pass
* Computes X * W + B
* X - input row vector
* W - weights matrix
* B - bias row wector
*/
output_vector forward(const input_vector& x)
{
/*
* Check for input size mismatch
*/
assert(x.size() == weights[0].size());
/*
* Layer output is dot product of input with weights
*/
output_vector activation;
transform(weights.begin(), weights.end(), bias.begin(), activation.begin(),
[x, this](const input_vector& w, T bias)
{
T val = inner_product(x.begin(), x.end(), w.begin(), 0.0);
if (use_bias)
{
val += bias;
}
return val;
}
);
return activation;
}
/*
* Dense layer backward pass
*/
input_vector backward(const input_vector& input, const output_vector grad)
{
/*
* Weight update according to SGD algorithm with momentum = 0.0 is:
* w = w - learning_rate * d_loss/dw
*
* For simplicity assume learning_rate = 1.0
*
* d_loss/dw = dloss/dy * dy/dw
* d_loss/dbias = dloss/dy * dy/dbias
*
* dloss/dy is input gradient grad
*
* dy/dw is :
* y = w[0]*x[0] + w[1] * x[1] +... + w[n] * x[n] + bias
* dy/dw[i] = x[i]
*
* dy/dbias is :
* dy/dbias = 1
*
* For clarity we:
* assume learning_rate = 1.0
* first compute dw
* second update weights by subtracting dw
*/
/*
* Compute backpropagated gradient
*/
vector<output_vector> weights_transposed;
weights_transposed.resize(num_inputs);
for (int i = 0; i < num_inputs; i++)
{
for (int j = 0; j < num_outputs; j++)
{
weights_transposed[i][j] = weights[j][i];
}
}
input_vector ret;
transform(weights_transposed.begin(), weights_transposed.end(), ret.begin(),
[grad](input_vector& w)
{
T val = inner_product(w.begin(), w.end(), grad.begin(), 0.0);
return val;
});
/*
* compute dw
* dw = outer(x, grad)
*/
vector<input_vector> dw;
for (auto grad_i: grad)
{
auto row = input;
for_each(row.begin(), row.end(), [grad_i](T &xi){ xi *= grad_i;});
dw.push_back(row);
}
/*
* compute w = w - dw
* assume learning rate = 1.0
*/
transform(weights.begin(), weights.end(), dw.begin(), weights.begin(),
[](input_vector& left, input_vector& right)
{
transform(left.begin(), left.end(), right.begin(),
left.begin(), minus<T>());
return left;
});
if (use_bias)
{
/*
* compute bias = bias - grad
* assume learning rate = 1.0
*/
transform(bias.begin(), bias.end(), grad.begin(), bias.begin(),
[](const T& bias_i, const T& grad_i)
{
return bias_i - grad_i;
});
}
return ret;
}
/*
* Helper function to convert Dense layer to string
* Used for printing the layer weights and biases
*/
operator std::string() const
{
std::ostringstream ret;
ret.precision(7);
/*
* output weights
*/
ret << "weights:" << std::endl;
for (int y=0; y < weights[0].size(); y++)
{
for (int x=0; x < weights.size(); x++)
{
if (weights[x][y] >= 0)
ret << " ";
ret << std::fixed << weights[x][y] << " ";
}
ret << std::endl;
}
if (use_bias)
{
/*
* output biases
*/
ret << "bias:" << std::endl;
for (auto b: bias)
{
if (b >= 0)
ret << " ";
ret << std::fixed << b << " ";
}
ret << std::endl;
}
return ret.str();
}
/*
* Helper function to cout Dense layer object
*/
friend ostream& operator<<(ostream& os, const Dense& dense)
{
os << (string)dense;
return os;
}
};Sigmoid layer class template includes forward() and backward() functions. forward() computes sigmoid(x) = 1 / (1 + exp(-x)) backward() computes mutiple of gradient and sigmoid derivative wrt input.
/*
* Sigmoid layer class template
*/
template<size_t num_inputs, typename T = float>
struct Sigmoid
{
typedef array<T, num_inputs> input_vector;
static input_vector forward(const input_vector& y)
{
input_vector ret;
transform(y.begin(), y.end(), ret.begin(),
[](const T& yi)
{
T out = 1.0 / (1.0 + expf(-yi));
return out;
});
return ret;
}
static input_vector backward(const input_vector& y, const input_vector grad)
{
input_vector ret;
transform(y.begin(), y.end(), grad.begin(), ret.begin(),
[](const T& y_i, const T& grad_i)
{
T s = 1.0 / (1.0 + expf(-y_i));
T out = grad_i * s * (1 - s);
return out;
});
return ret;
}
};Softmax layer implementation includes forward and backward path. Forward path computes Softmax(x) = exp(x) / sum (exp(x)) Backward path is dot product of input gradient and Softmax Jacobian as per math section above.
/*
* Softmax layer class template
*/
template<size_t num_inputs, typename T = float>
struct Softmax
{
typedef array<T, num_inputs> input_vector;
/*
* Softmax forward function
*/
static input_vector forward(const input_vector& x)
{
input_vector y;
/*
* compute exp(x_i) / sum(exp(x_i), i=1..N)
*/
transform(x.begin(), x.end(), y.begin(),
[](const T& yi)
{
T out = expf(yi);
return out;
});
T sum = accumulate(y.begin(), y.end(), static_cast<T>(0));
for_each(y.begin(), y.end(), [sum](T &yi){ yi /= sum;});
return y;
}
/*
* Softmax backward function
*/
static input_vector backward(const input_vector& x, const input_vector& grad_inp)
{
input_vector grad_out;
input_vector y;
vector<input_vector> J;
y = forward(x);
int s_i_j = 0;
/*
* Compute Jacobian of Softmax
*/
for (auto y_i: y)
{
auto row = y;
for_each(row.begin(), row.end(), [y_i](T& y_j){ y_j = -y_i * y_j;});
row[s_i_j] += y_i;
s_i_j++;
J.push_back(row);
}
/*
* Compute dot product of gradient and Softmax Jacobian
*/
transform(J.begin(), J.end(), grad_out.begin(),
[grad_inp](const input_vector& j)
{
T val = inner_product(j.begin(), j.end(), grad_inp.begin(), 0.0);
return val;
}
);
return grad_out;
}
};Mean Squared Error class will need it’s own forward and backward functions.
/*
* Mean Squared Error loss class
* Parameters:
* num_inputs: number of inputs to MSE function.
* T: input type, float by defaut.
*/
template<size_t num_inputs, typename T = float>
struct MSE
{
/*
* Forward pass computes MSE loss for inputs y (label) and yhat (predicted)
*/
static T forward(const array<T, num_inputs>& y, const array<T, num_inputs>& yhat)
{
T loss = transform_reduce(y.begin(), y.end(), yhat.begin(), 0.0, plus<T>(),
[](const T& left, const T& right)
{
return (left - right) * (left - right);
}
);
return loss / num_inputs;
}
/*
* Backward pass computes dloss/dy for inputs y (label) and yhat (predicted)
*
* loss = sum((yhat[i] - y[i])^2) / N
* i=0...N-1
* where N is number of inputs
*
* d_loss/dy[i] = 2 * (yhat[i] - y[i]) * (-1) / N
* d_loss/dy[i] = 2 * (y[i] - yhat[i]) / N
*
*/
static array<T, num_inputs> backward(const array<T, num_inputs>& y,
const array<T, num_inputs>& yhat)
{
array<T, num_inputs> de_dy;
transform(y.begin(), y.end(), yhat.begin(), de_dy.begin(),
[](const T& left, const T& right)
{
return 2 * (right - left) / num_inputs;
}
);
return de_dy;
}
};Finally, in the main function, we’ll declare input x and expecetd output y_true arrays, containing the same values as in out Python example. Then we’ll compute forward and backward passes, and print the network output and updated weights.
int main(void)
{
const int num_inputs = 2;
const int num_outputs = 2;
const int num_iterations = 1000;
array<float, num_inputs> x = {-1.0, 0.0};
array<float, num_outputs> ytrue = {1.0, 0.0};
array<float, num_outputs> biases_init = {1.0, 2.0};
array<array<float, num_outputs>, num_outputs> weights_init =
{
{
{1.0, 3.0},
{2.0, 2.0}
}
};
/*
* Create dense layers and MSE loss
*/
Dense<num_inputs, num_outputs, float, const_initializer<const_one>, const_initializer<const_two> > dense1(true);
Sigmoid<num_outputs> sigmoid;
Dense<num_outputs, num_outputs, float, const_initializer<const_one> > dense2(weights_init, biases_init);
Softmax<num_outputs> softmax;
MSE<num_outputs> mse_loss;
/*
* Compute Dense layer output y for input x
*/
auto y1 = dense1.forward(x);
auto y2 = sigmoid.forward(y1);
auto y3 = dense2.forward(y2);
auto y4 = softmax.forward(y3);
/*
* Copute MSE loss for output y4 and label ytrue
*/
auto loss = mse_loss.forward(ytrue, y4);
/*
* Benchmark Dense layer inference latency
*/
auto ts = high_resolution_clock::now();
for (auto iter = 0; iter < num_iterations; iter++)
{
//y1 = dense1.forward(x);
}
auto te = high_resolution_clock::now();
auto dt_us = (float)duration_cast<microseconds>(te - ts).count() / num_iterations;
/*
* Print DNN input x
*/
printf("input x=");
for_each(x.begin(), x.end(), print_fn);
printf("\n");
/*
* Print DNN output y and expected output yhat
*/
printf("output y=");
for_each(y4.begin(), y4.end(), print_fn);
printf("\n");
printf("expected output ytrue=");
for_each(ytrue.begin(), ytrue.end(), print_fn);
printf("\n");
/*
* Print loss
*/
printf("loss: %f\n", loss);
/*
* Back propagate loss
*/
auto dloss_dy4 = mse_loss.backward(ytrue, y4);
auto dy4_dy3 = softmax.backward(y3, dloss_dy4);
auto dy3_dy2 = dense2.backward(y2, dy4_dy3);
auto dy2_dy1 = sigmoid.backward(y1, dy3_dy2);
dense1.backward(x, dy2_dy1);
printf("input x=");
for_each(x.begin(), x.end(), print_fn);
printf("\n");
/*
* print dloss/dy
*/
printf("dloss/dy: ");
for_each(dloss_dy4.begin(), dloss_dy4.end(), print_fn);
printf("\n");
/*
* Print updated Dense layer weights
*/
printf("updated dense 1 layer weights:\n%s", ((string)dense1).c_str());
printf("updated dense 2 layer weights:\n%s", ((string)dense2).c_str());
/*
* Print average latency
*/
printf("time dt=%f usec\n", dt_us);
return 0;
}After compiling and running C++ example:
$ g++ -o dense5 -std=c++2a dense5.cpp && ./dense5
input x=-1.0000000 0.0000000
output y=0.2689414 0.7310586
expected output ytrue=1.0000000 0.0000000
loss: 0.534447
input x=-1.0000000 0.0000000
dloss/dy: -0.7310586 0.7310586
updated dense 1 layer weights:
weights:
1.0565200 0.9434800
1.0000000 1.0000000
bias:
1.9434800 2.0565200
updated dense 2 layer weights:
weights:
1.2101572 1.7898428
3.2101572 1.7898428
bias:
1.2874696 1.7125304
time dt=0.001000 usecAs one can verify, forward path output of the C++ implementation matches the Python code. Also, weights and biases of dense layers after backpropagation match in Python and C++ code.
Previous example can be found at “Dense layer with backpropagation and sigmoid activation in C++”
Python source code for this example is at dense5.py
C++ implementation is at dense5.cpp